DNA replication, also known as semi-conservative replication, is the process by which DNA is doubled. This is an important process taking place within the dividing cell.
In this article, we shall discuss the structure of DNA, the steps involved in DNA replication (initiation, elongation and termination) and the clinical consequences that can occur when this process goes wrong.
DNA Structure
DNA is made up of millions of nucleotides. These are molecules composed of a deoxyribose sugar, with a phosphate and a base (or nucleobase) attached to it. These nucleotides are attached to each other in strands via phosphodiester bonds to form a ‘sugar-phosphate backbone’. The bond formed is between the third carbon atom on the deoxyribose sugar of one nucleotide (known as the 3’) and the fifth carbon atom of another sugar on the next nucleotide (known as the 5’).
N.B: 3′ is pronounced ‘three prime’ and 5′ is pronounced ‘five prime’.
There are two strands of DNA, which run in opposite (antiparallel) directions to each other. These strands are attached to each other throughout their lengths via the bases on each nucleotide.
There are 4 different bases associated with DNA: Cytosine, Guanine, Adenine, and Thymine. In normal DNA strands, cytosine binds to guanine, and adenine binds to thymine. When bound together, the two strands form a double helix structure.
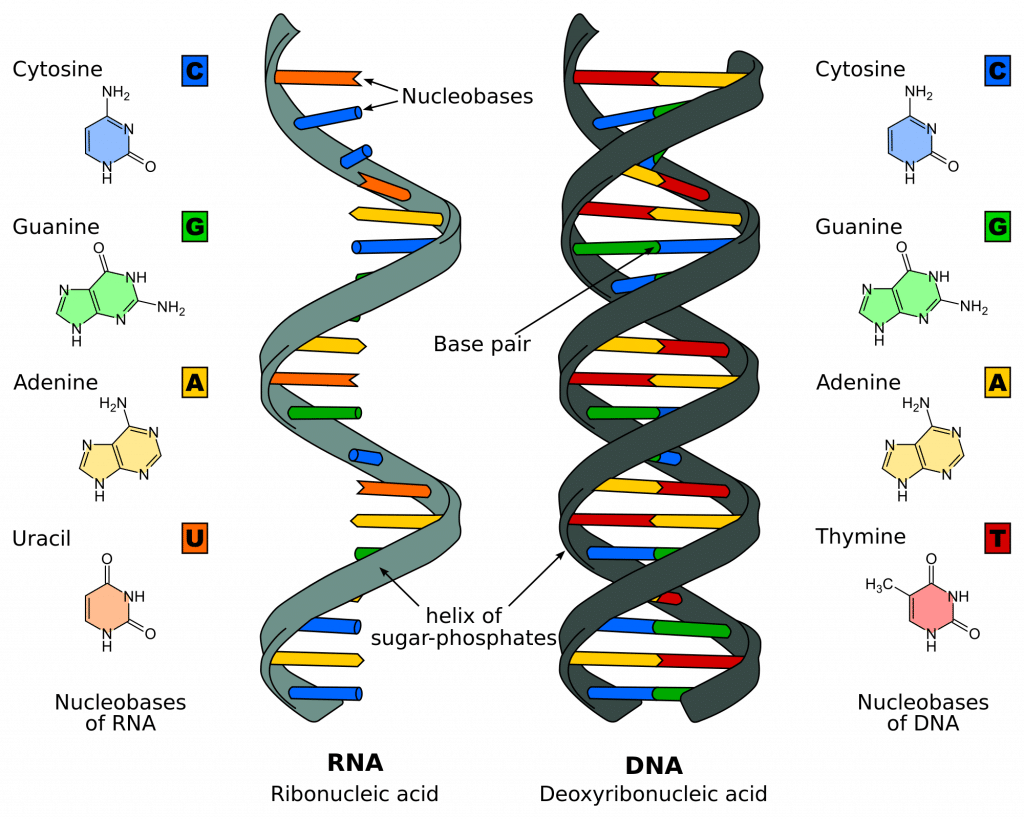
Fig 1 – Diagram showing the difference between the structure of DNA and RNA
Stages of DNA replication
DNA replication can be thought of in three stages: initiation, elongation and termination
Initiation
DNA synthesis is initiated at particular points within the DNA strand known as ‘origins’, which have specific coding regions. These origins are targeted by initiator proteins, which go on to recruit more proteins that help aid the replication process, forming a replication complex around the DNA origin. Multiple origin sites exist within the DNA’s structure; when replication of DNA begins, these sites are referred to as replication forks.
Within the replication complex is the DNA helicase. This enzyme unwinds the double helix and exposes each of the two strands so that they can be used as a template for replication. It does this by hydrolysing the ATP used to form the bonds between the nucleobases, thereby breaking the bond holding the two strands together.
DNA primase is another enzyme that is important in DNA replication. It synthesises a small RNA primer, which acts as a ‘kick-starter’ for DNA polymerase. This enzyme is ultimately responsible for the creation and expansion of new strands of DNA.
Elongation
Once DNA Polymerase has attached to the two unzipped strands of DNA (i.e. the template strands), it is able to start synthesising new strands of DNA to match the templates. DNA polymerase is only able to extend the primer by adding free nucleotides to the 3’ end.
One of the template strands is read in a 3’ to 5’ direction, therefore the new strand will be formed in a 5’ to 3’ direction. This newly formed strand is referred to as the leading strand. Along the leading strand, DNA primase only needs to synthesise an RNA primer once, at the beginning, to initiate DNA polymerase. This is because DNA polymerase is able to extend the new DNA strand by reading the template 3′ to 5′, synthesising in a 5′ to 3′ direction as noted above.
However, the other template strand (the lagging strand) is antiparallel and is therefore read in a 5’ to 3’ direction. Continuous DNA synthesis, as in the leading strand, would need to be in the 3′ to 5′ direction, which is impossible as DNA polymerase cannot add bases to the 5′ end. Instead, as the helix unwinds, RNA primers are added to the newly exposed bases on the lagging strand and DNA synthesis occurs in fragments, but still in the 5′ to 3′ direction as before. These fragments are known as Okazaki fragments.
Termination
The process of expanding the new DNA strands continues until there is either no more DNA template strand left to replicate (i.e. at the end of the chromosome) or two replication forks meet and subsequently terminate. The meeting of two replication forks is not regulated and happens randomly along the course of the chromosome.
Once DNA synthesis has finished, the newly synthesised strands are bound and stabilised. For the lagging strand, two enzymes are needed to achieve this stabilisation: RNAase H removes the RNA primer at the beginning of each Okazaki fragment, and DNA ligase joins these fragments together to create one complete strand.
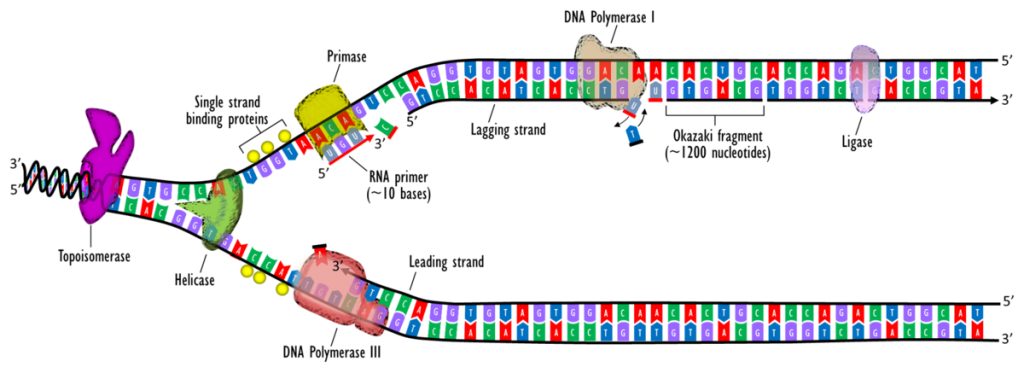
Fig 2 – Diagrammatic representation of DNA replication
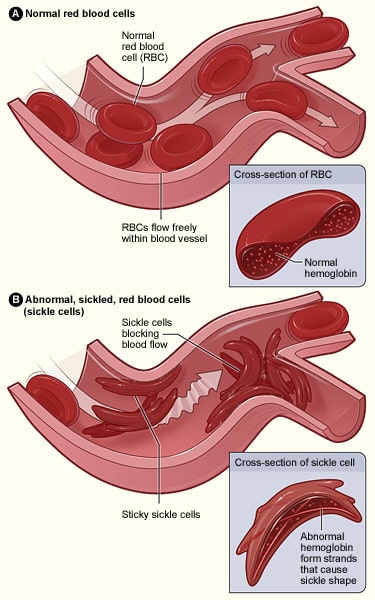
Clinical Relevance – Sickle Cell Anaemia
Sickle cell anaemia is an autosomal recessive condition which is caused by a single base substitution, in which one single base is changed for another. In some cases, single base substitutions can result in a ‘silent mutation‘ in which the overall gene is not affected, however, in diseases such as sickle cell anaemia, it results in the strand coding for a different protein.
In this case, an adenine base is swapped for a thymine base in one of the genes coding for haemoglobin; this results in glutamic acid being replaced by valine. When this is being transcribed into a polypeptide chain, the properties it possesses are radically changed, as glutamic acid is hydrophilic, whereas valine is hydrophobic. This hydrophobic region results in haemoglobin having an abnormal structure that can cause blockages of capillaries, leading to ischaemia and potentially necrosis of tissues and organs – this is known as a vaso-occlusive crisis.
These crises are typically managed with analgesia, including opioids and NSAIDs depending on the severity. Red blood cell transfusions may be required in emergencies, for example, if the blockage occurs in the lungs.